Imaging-Based DNA Sequencing: Past, Present & Future [Part 2]
How image analysis and computational tools have taken genome sequencing from years to minutes; from population to personal
This is Part Two of Imaging-Based DNA Sequencing: Past, Present & Future. If you haven’t read it, you can find it here.
Third-generation sequencing: Methods proliferate
Today, Illumina is the clear market leader of NGS, and as Third-Generation Sequencing (TGS) arrive, there is another wave of new technologies on their way with hopes to improve on the short read lengths of Second Generation technologies. NGS technologies all require large genome DNA to be fragmented and amplified due to the limitations in read lengths. This fragmentation and amplification has significant consequences, including amplification bias of repetitive DNA units by PCR and the need for computational models to align the DNA fragments. There is not always sufficient fragment overlap to accurately align segments of DNA, which makes de novo, or discovery sequencing, challenging with NGS techniques. TGS is most easily defined as sequencing without amplification, and typically is characterized by much longer read-lengths than for NGS. There have been several approaches to accomplishing long read lengths in sequencing.
PacBio led the charge with their zero-mode waveguide technology. The technology used nano holes to tether a single DNA polymerase, which allowed real-time observation of each addition of a fluorescently labelled nucleotide to the chain. This process was named Single Molecule Real Time (SMRT) sequencing. And additional companies came on board with PacBio, including the Oxford Nanopore device, which doesn’t use image analysis at all but rather monitors electrical activity.
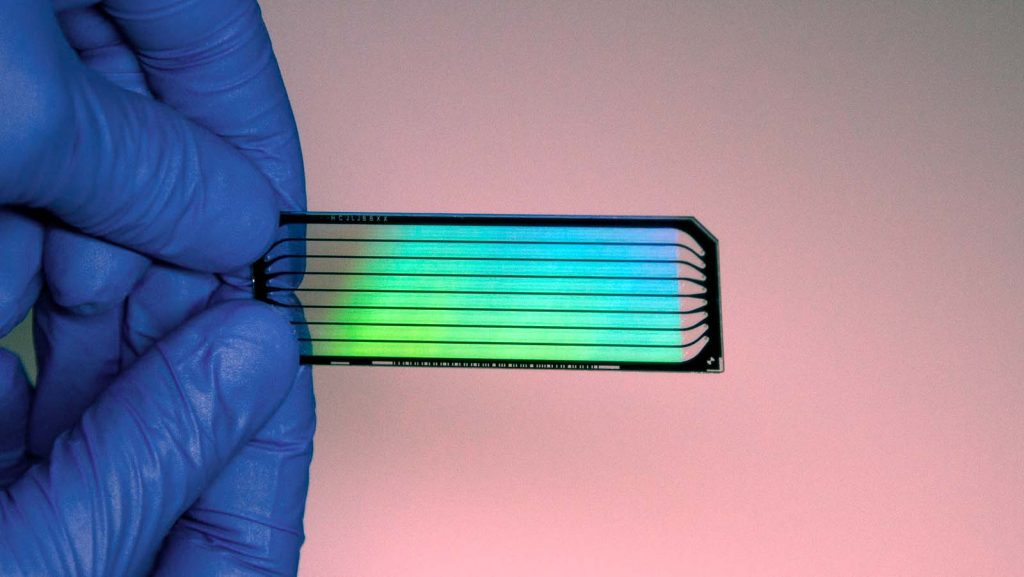
SMRT sequencing can produce read lengths of 10-20 thousand base pairs. Although the error rate was higher than Illumina’s machines, there was reduced sequencing bias, and the sequencing data was high quality, despite the limitations. Updates to this methodology led PacBio to release a High Fidelity (HiFi) machine in 2019 with long reads at the same accuracy of the short reads produced by Illumina. That same year, PacBio released a new machine Sequel II with new chemicals and a novel ZMW nanowell smartcell that can produce up to 4M HiFi reads with 99.999% accuracy, and dramatically reduces the sequencing cost.
TGS platforms produce relatively noisy data which has led to significant reliance on computational tools to get reliable results. PacBio’s FALCON uses Hierarchical Genome Assembly Process (HGAP) technology. This software takes the longest read produced and uses it to align shorter reads; a consensus read is created from all of the reads based on computational algorithms developed in the mid-nineties (de Bruijn Graphs). Canu is another alternative software developed for this purpose; it creates sequences through the overlap layout-consensus of long reads. One challenge for TGS is that the time for assembly based on computational methods is becoming longer than the time for sequencing and the same can be said about the computational costs.
In the Third-Generation landscape, Illumina continues to fight for market shares with its NovaSeq platform that can generate up to 20B bp per run. However, this platform is limited to short reads less than 250bp and requires use of a suite of computational tools for sequence alignment. Illumina itself has its own suite of software tools for use with their sequencers. Everything from lab automation tools to AI-based analytical tools and a platform called DRAGEN that claims to process the entire human genome in 25 minutes! That’s certainly dramatically faster than 13 years. The future of sequencing is likely a combination of NGS and TGS techniques, taking advantage of the strengths of each system to create complete, accurate genome sequences – at the individual level.
Looking forward: More data, more imaging, more personal
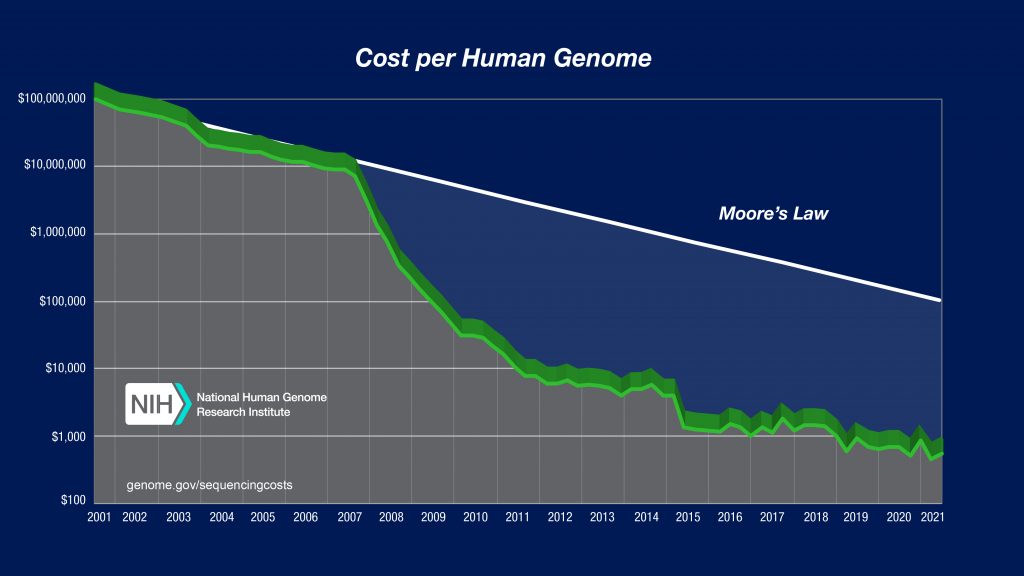
The era of personalized medicine is here and is expected to become more accessible to the individual. However, with that access a familiar challenge arises, how do we save all of this data in a retrievable and safe way? This is such a challenge for researchers that tools have been developed to help labs estimate how much storage they need and what types of storage (e.g. cloud, server) and the associated costs. Raw imaging data can occupy a huge amount of storage space, which can be expensive to manage. In response, many technologies are no longer storing the raw image files acquired during sequencing, and instead compress a whole genome data into a relatively more manageable 100GB. That’s still a lot of data, but it also seems remarkably small to describe the recipe for an entire human being.
The arrival of personalized medicine is powerful, it allows early cancer detection, genetic screening, improved diagnostics and personalized therapeutics. While this level of personalization has the potential to lead to better medical outcomes, it comes with a host of challenges. 100GB for a genome seems rather reasonable, until you imagine millions of individuals mapping their genes, the storage requirements start to become staggeringly large. In addition, data protection should be on everyone’s mind as we begin generating individual sequencing. The advent of genealogy companies like 23andme have highlighted an interesting question of what ownership we have of our heredity and what resources we have to protect our genetic data.
Some scientists predict that moving from population-based genomics to single-cell genomics will be a critical step in the precision medicine journey. Single-cell genomics can allow three-dimensional imaging of a cell, aided by super-resolution microscopy – to visualize cell fate in living tissue. This, combined with the ability to manipulate the genome using CRISPR can lead to discovery of gene mutation pathways and prediction of cell behavior, which in turn could lead to more targeted therapies and cures. The 2030 Bold Predictions for human genomics put forth by the National Human Genome Research Institute (NHGRI) predicts that genome sequencing will become a routine part of medicine, like getting blood drawn.
Imaging and image analysis tools will continue to be critical in the development of personalized medicine. The current approach to genome sequencing deals with the genome in a static state – but an individual genome is not static, our bodies are full of dividing cells and mutations occur over time. To this end, a multitude of techniques are being developed to study genome dynamics in real time. Use of rapidly evolving light sheet microscopy, and other super-resolution techniques like stochastic optical reconstruction microscopy (STORM), photo-activated localized microscopy (PALM) and stimulation emission depletion (STED) microcopy will allow better understanding of the relationship between structure and function at the cellular level.
NGS, TGS, novel imaging techniques, and advancements in data analysis tools will be combined to create genomic information at our fingertips, or as the NHGRI 2030 predictions state, our personal genome will be “available on our smart phones in a user-friendly form.”


 Advancing genome sequencing at the speed of light
Advancing genome sequencing at the speed of light  Lock And Load: Shotgun Metagenomics and SARS-CoV-2
Lock And Load: Shotgun Metagenomics and SARS-CoV-2 