DNA Sequencing is Getting Faster, More Accurate and Mobile
The rapid maturation of DNA sequencing technology has given rise to an explosion of genomic data that could help fundamentally change healthcare and science.
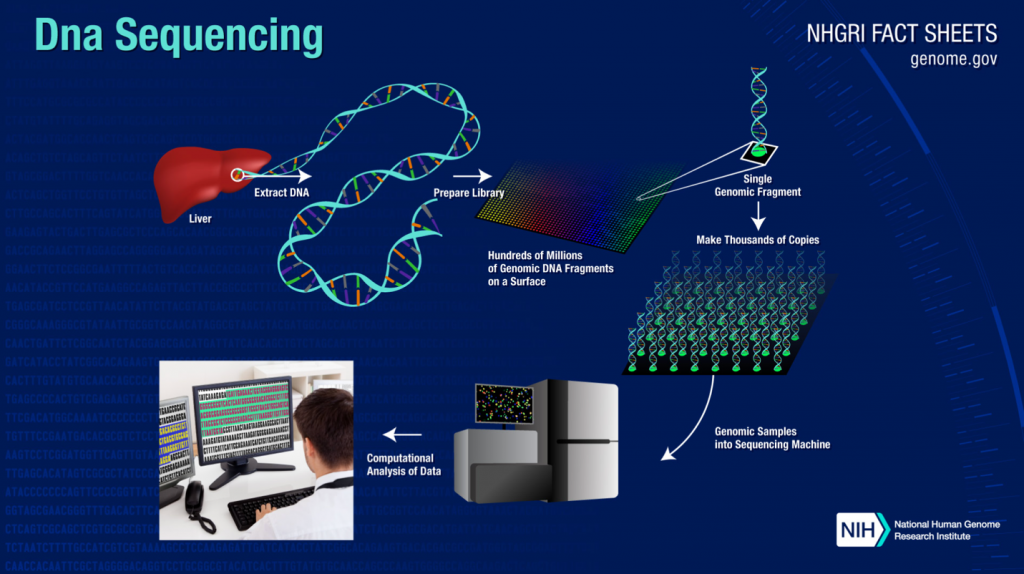
The human genome has 19,000 to 22,000 genes, all combining to tell the story of our bodies. DNA and RNA and full pairs of chromosomes hold a lot of data, to the point that it used to take supercomputers to do the heavy lifting of DNA sequencing. And locating a specific gene could take days or weeks, if it could be found at all.
The march of technology has made DNA sequencing faster, more reliable, and accurate. DNA sequencing can now be done with a handheld device paired with a smartphone. And researchers are now able to isolate and analyze specific genes without having to perform the cumbersome process of sequencing an entire strand of DNA.
The first full DNA genome ever sequenced was the humble PhiX174, the single-strand virus that infects E.coli. Developed by British biochemist Fred Sanger and colleagues in 1977, the “dideoxy chain-termination method” for sequencing DNA molecules—called the “Sanger method”—dominated the field of gene sequencing for the next 30 years and was eventually used to sequence the entire human genome.
Completed in 2003, the Human Genome Project took 13 years to complete and cost about $3 billion. With today’s technology, the project could be completed in about a day at a cost closer to $1,000. And with an app like iGenomics, it could eventually be done with a smartphone.
Even with the advance in technology, the human genome has three billion data points. Finding a specific gene—such as a mutated gene that causes cancer—has historically been very difficult, if it could be accomplished at all. But researchers developed a method in 2020 that can help isolate regions of genes for rapid analysis, promising quicker and more precise diagnosis in the future.
From the Sanger Method to Isolating Specific Genes
While the Sanger method dominated the field for three decades, it was very slow and imprecise. After the Human Genome Project concluded, scientists began releasing new kinds of DNA sequencing methods that are collectively known as Next Generation Sequencing (NGS).
Next Generation Sequencing techniques have led to an explosion of data in genomics. Researchers have been building databases of rare and infectious diseases, cancers, and psychiatric disorders. The 100,000 Genomes Project was an initiative from the United Kingdom that began in 2012 to sequence the genomes of 100,000 people with rare diseases or cancers to build the most comprehensive genomics database of its kind ever created. The PyscheENCODE project compiled data from 2,000 human brains to “form the most complete picture of how regions that regulate DNA expression can influence the brain and its function.” These kinds of databases couldn’t have been completed with the Sanger method.
One NGS method (or third generation, depending on whom you ask) that has gained traction within the last decade is called “nanopore sequencing.” A nanopore is what it sounds like, a pore of nano size. According to Oxford Nanopore Technologies, the method “enables direct, real-time analysis of long DNA or RNA fragments. It works by monitoring changes to an electrical current as nucleic acids are passed through a protein nanopore. The resulting signal is decoded to provide the specific DNA or RNA sequence.” Nanopore sequencing has promised to offer low-cost genotyping, rapid processing of samples with results in real time, and high mobility. The method has been used for the identification of viral pathogens, human or plant genome sequencing, and monitoring of antibiotic resistance, among other uses.
In November 2020, scientists led by a team from the University of Washington announced a breakthrough using a nanopore technique called Targeted Long Read Sequencing (T-LRS) that allowed them to rapidly isolate specific regions of gene. In a trial of 33 people with known genetic disorders, the method—called Read Until—proved to be more accurate than either PCR or Cas-9 techniques (terms you might recognize from the news of testing methods for COVID-19).
The researchers describe how the method works:
“We applied the adaptive sequencing mode known as Read Until using the ReadFish software package, which allowed us to dynamically select target regions for sequencing. In this mode, software analyzes the signal after a DNA molecule enters a pore to determine whether that molecule lies within a specified genomic region of interest. If it does, the pore continues to sequence the molecule; if not, the DNA molecule is ejected from the pore.”
The study was completed using a nanopore sequencing device called GridION from Oxford Nanopore that is about the size of a desktop printer.
Imaging at the Molecular Level
Scientists have continually gotten better at reading and sequencing DNA at its most minute scales.
Researchers out of the University of Tokyo published a paper in December 2020 that described a unique method for spatial decoding of DNA barcodes at the resolution of a single molecule. DNA barcodes are specific short sections of DNA from a gene that is often used to identify species, like how a barcode is used to identify a product at a supermarket.
The researchers repurposed a method used from a Heliscope machine, from the now-defunct Helicose Biosciences company. Called “single molecule fluorescent sequencing,” the method works by imaging a single fluorophore molecule (a fluorophore is chemical compound that can re-emit light after being excited) without having to replicate the DNA through amplification methods, which could lead to bias in the results.
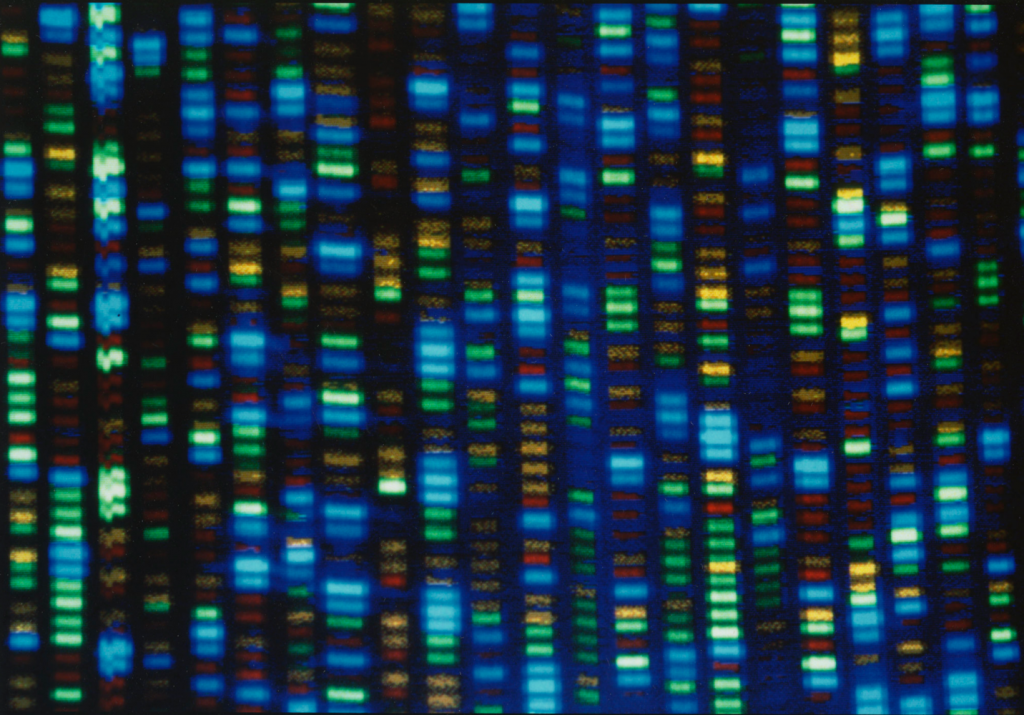
Essentially, the team used fluorescence microscopy to create a novel method of DNA sequencing at the molecular level by repurposing and improving on a method that had gone out of favor among researchers. The method improves upon existing practices because it does not require a large sample of DNA, does not require replication of the sample through amplification, and can analyze samples at the single molecule level.
Sequencing and Analysis, Anywhere
The history of DNA sequencing has been set in laboratory environments. The instruments were large and cumbersome, sitting on benches and desks alongside equally large computers. Moore’s Law, like it has with everything else computing, has changed that. The smartphones in our pockets are more powerful than the advanced laptops from 10 years ago. It was thus inevitable for DNA sequencing to make its way out of the lab and into the field.
Oxford Nanopore Technologies has been on the forefront of the miniaturization of DNA sequencing. Its MinION sequencer is the size of a thick harmonica and weighs 450 grams. And yet, analysis of the sequence would often still need to be done on a laptop or desktop computer, limiting its benefits when used in field testing.

A project called iGenomics hopes to change that. iGenomics is an iPhone app that was designed to complement the DNA sequencer devices made by Oxford Nanopore. By removing the need for a large laptop or cloud computer to analyze the data generation by devices like the MinION, iGenomics hopes to free DNA sequencing to go . . . anywhere.
DNA sequencing aboard the International Space Station is a distinct possibility. Or testing for virus variants such as Zika or COVID-19 in remote locations of the world.
“The true novelty of this application is not in the algorithms used but rather how they have been implemented in a mobile environment,” the creators of iGenomics wrote in a research paper published in December 2020. “An important use case of iGenomics could be a researcher with limited computational resources sequencing complementary DNA (cDNA) of a coronavirus sample, loading and aligning the cDNA reads with iGenomics, and getting a first analysis of the coronavirus mutations within a few seconds.”
Faster, More Accurate, and Mobile
The rapid evolution of DNA sequencing methods combined with the power of big data and mobility promises a future of genomic breakthroughs that few could have imagined a few decades ago. Soon we will likely be able to quickly diagnose a form of cancer, sequence its genetics, then use artificial intelligence to build medicines to treat it, unique to the individual patient. Genetic variants of contagious diseases can be identified quicker, wherever they are found. The unique DNA barcodes of plant and animal species can be studied in the field, fundamentally changing the nature of bioscience.


 Genomics takes its place in the race against COVID-19
Genomics takes its place in the race against COVID-19  Advancing genome sequencing at the speed of light
Advancing genome sequencing at the speed of light 